In the last post we described the theory necessary to perform causal inference in observational data, but we left unanswered some questions on how to estimate the propensity score function. We made this choice, because the details on the best way to estimated such function are not that simple and needed a exclusive post, so the explanation would be more consistent.
PS Model Should Not Predict The Treatment Variable As Well As Possible
There is a common misperception about how to estimate the PS model, when the analyst learning about causal inference already has some experience using machine learning algorithms to solve prediction problems. Hernán in his forthcoming book bring this problem to our attention:
“Unfortunately, statistics courses and textbooks have not always made a sharp difference between causal inference and prediction. […] Specifically, the application of predictive algorithms to causal inference models may result in inflated variances […]. One of the reasons for variance inflation is the widespread, but mistaken, belief that propensity models should predict treatment T as well as possible. Propensity models do not need to predict treatment very well. They just need to include the covariates that guarantee exchangeability.” – Chapter 15, Section 15.5 – Hernán MA, Robins JM (2016). Causal Inference. Boca Raton: Chapman & Hall/CRC, forthcoming.
This lights up a red flag for us as we intend to use machine learning algorithms to do this estimation. The purpose of the propensity score model is not to separate the best way possible both treatment classes, but to best estimate the treatment assignment probability of each observation. Such statement forces us to ask an important question: which error measurement best suits such purpose? Besides that, Hernán also says that including the correct covariates to estimate propensity score model is enough to avoid misspecification. Well, if we limit our statistical algorithms to GLM with no regularization, such statement is valid, once it does not require search for the best hyperparameters. However, again, we intend to use machine learning algorithms to perform such task. So, first we need to establish a novel criteria, different from logloss and AUC error metrics, to measure the performance of models for this specific purpose, before we can start doing some programming.
Measuring Treatment Assignment Probability Error: Heterogeneity Metric
Now, we need to understand what implies finding the best estimate for the treatment assignment probability of each observation. Suppose we specify the statistical model perfectly and we use the yielded probabilities to apply the process to create a blockage between T and its parents covariates. In this way, we would establish the exchangeability condition between treatment and control group. Which means that the distribution of all covariates parents of T are equal or pretty close, i.e. homogeneous, between treatment and control group. Therefore, to solve our problem we need to define a procedure to measure the heterogeneity between both groups for the covariates in sets 1 and 2 and use it to find the estimated model that minimizes the measured heterogeneity.
Our heterogeneity metric was inspired in the ideas behind the R package called twang and it is based on two homogeneity tests: Kolmogorov-Smirnov (KS) and Chi-squared (X2). The latter for categorical variables and the former for continuous ones. The Heterogeneity Metric is calculated by applying the correct homogeneity test to compare the distribution of a covariate between the treatment and the control groups. These covariates are the ones in sets 1 e 2, described in the previous post. Each homogeneity test varies from 0 to 1, where 0 represents minimum and 1 maximum heterogeneity. Next, we sum each covariate’s result and obtain the Heterogeneity Metric. Summing up, Heterogeneity Metric = Σ (KS or X2) for each covariate parent of treatment variable.
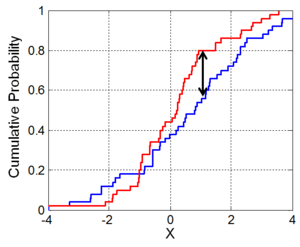
To give a better understanding about the homogeneity tests, we’ll show an short example of both. To understand how the Kolmogorov-Smirnov (KS) homogeneity test is calculated, we just need to take a look in the next figure. We sort the observation by the value of the covariate X and draw in a chart its cumulative distribution for the treatment (blue) and the control (red) group. To make them comparable, we need to normalize the sum of weights to 1 inside each specific group. The KS value is the biggest vertical distance between the two curves. Here indicated by the two headed arrow. As you can notice, its value is always inside the interval [0, 1]. As a sort operation is required and the datasets can be really big, we used the R package, data.table, to order the observations, once it uses a very fast and very memory efficient implementation of the radix sort algorithm.
The other test is Chi-squared (X2) test of homogeneity that is applied to a single categorical variable from two different populations (control and treatment) to determine whether frequency counts are distributed identically across these different populations. Let’s use a simple example to show how it is calculated: in a study of the television viewing habits of children, a developmental psychologist selects a random sample of 300 first graders – 100 boys and 200 girls. Each child is asked which of the following TV programs they like best: The Lone Ranger, Sesame Street, or The Simpsons.
| Observed | Viewing Preferences | Row total | ||
|
Lone Ranger |
Sesame Street | The Simpsons | ||
| Boys | 50 | 30 | 20 | 100 |
| Girls | 50 | 80 | 70 | 200 |
| Column total | 100 | 110 | 90 | 300 |
How homogeneous is the boys’ preferences for these TV programs when compared to the girls’ ones? To obtain the heterogeneity measure, first we need to normalize the counts, so the final result will be inside interval [0,1].
| Observed and Normalized | Viewing Preferences | Row total | ||
|
Lone Ranger |
Sesame Street | The Simpsons | ||
| Boys | 0.1667 | 0.1000 | 0.0667 | 0.3333 |
| Girls | 0.1667 | 0.2667 | 0.2333 | 0.6667 |
| Column total | 0.3333 | 0.3667 | 0.3000 | 1.000 |
Second, we calculate the expected count for each cell: Er,c = (nr * nc) / n where Er,c is the expected frequency count for population r at level c of the categorical variable, nr is the total number of observations from population r, nc is the total number of observations at treatment level c, and n is the total sample size.
| Expected | Viewing Preferences | ||
| Lone Ranger | Sesame Street | The Simpsons | |
| Boys | 0.1111 | 0.1222 | 0.1000 |
| Girls | 0.2222 | 0.2444 | 0.2000 |
Third, we calculate the chi-squared for each cell: X2 = Σ [ (Or,c – Er,c)^2 / Er,c ] where Or,c is the normalized observed frequency count in population r for level c of the categorical variable, and Er,c is the expected frequency count in population r for level c of the categorical variable.
| Partial X2 | Viewing Preferences | ||
| Lone Ranger | Sesame Street | The Simpsons | |
| Boys | 0.0278 | 0.0040 | 0.0111 |
| Girls | 0.0139 | 0.0020 | 0.0056 |
And finally, we sum the chi-squared of all cells to find the total chi-squared: Total X2 = 0.0278 + 0.0139 + 0.0040 + 0.0020 + 0.0111 + 0.0056 = 0.0644. Which indicates that both preferences are relatively homogeneous and not that different. As a note, we implemented such algorithm using the R package data.table for a fast and memory efficient aggregation on big datasets.
You should have in mind, that we don’t want and don’t need to calculate the p-value for neither the homogeneity tests. We just obtain the statistics of the test for each covariate and add all of them to find the heterogeneity metric for the current weights.
Optimizing without using derivatives
Now that we have defined the metric, we need to establish how we’ll gonna use it with a machine learning engine. We know for sure that such metric is not defined in the engine itself, once such engines are normally used to maximize the prediction of labels, in the classification case, requiring very different error metrics. Therefore, we can only implement our algorithm using the engine to generate a model without performance evaluation and, externally to the engine, we calculate the respective error using a validation dataset. As you can imagine this can be very inefficient, once to find the best model we would need to test a huge amount of models consuming too much time and resources.
Therefore, we need to find a search algorithm that minimizes the heterogeneity metric based at least in one of the dimensions of the hyperparameters of the model, without the need of derivatives. The best way to achieve such requirements is analyzing the general chart: Error vs Model Complexity for training and validation datasets. In the next image, we notice that the train error, using a train dataset, is monotonic downward as the model’s complexity increases. However, the validation error, using a validation dataset, has a U shape, decreasing for the initial values of complexity and increasing for later ones. The vertical line that passes at the minimum value of the validation error separates the chart in two undesirable areas. In the left area, we have all models which the complexity underfits the data, i.e. it has high bias and low variance. And in the right one, which contains all models with complexity that overfits the data, we have low bias with high variance. The sweet spot is in the minimum error value for the validation dataset, where the balance between bias and variance is the best.
You may be wondering where is the test dataset and the cross-validation procedure to guarantee the model’s performance on new data. That is the good news, as this model just make sense for the current observational dataset, it just has to be the best model that represents the propensity score function for the data at hand, without overfitting it.
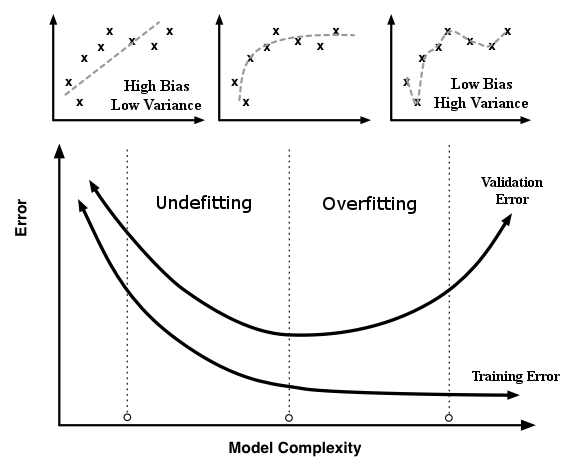
Error vs Model Complexity Chart
Now we need to define which parameter will represent the X axis of the chart. But first we need to define the 3 machine learning algorithms that we’ll gonna use in the demo (part 4 and 5) of this post series:
- GLM with Elastic Net Regularization, which has two hyperparameters: lambda and alpha. The selected parameter to represent the X axis is log10lambda. Some disadvantages of this algorithm is: it needs an trial and error search for the high order interaction terms, it requires one hot encoding for categorical variables and also standardization of numerical ones;
- GBM, which was first proposed to estimate PS function by (McCaffrey et al. 2004), has four hyperparameters: number of trees, max depth, learning rate and learning rate annealing. The selected parameter to represent the X axis is number of trees. Some advantages of this algorithm: it deals naturally with high order interaction terms, with categorical variables and it doesn’t need standardization of numerical variables.
- And of course, the GLM without Regularization, which is the default approach of vast majority of statisticians. Once, it doesn’t need a hyperparameters search, it is very simple to use and all this discussion about error measurement so far doesn’t apply to it. After estimating the model usually the analyst compares the mean of each covariate between both groups to have an idea how small is the final distribution discrepancy. If it still not small enough, she performs a trial and error search for covariates high order interaction terms trying to minimize the difference between the means. As it is well known and the most used algorithm, we’ll use it as our benchmark, so the heterogeneity metric calculated for it will just have the comparison purpose between its behaviour and the other algorithms’ one.
An reminder, all selected machine learning algorithms must support weight for each observation, as for each iteration new weights will be calculated and used in the next iteration to estimate a new statistical model.
So after defining the error metric and the function which needs to be minimized, the next step is to define how to execute the minimization search itself. As one of the requirements is minimization without using derivatives, we chose the R’s optimize method that is a combination of golden section search and successive parabolic interpolation to find the local minimum value of a function or the global minimum if such function is unimodal.
As the optimize method could call the function to be optimized with the same value more than once, it is a good idea to implement such function using a closure, to enable us to cache during a search all calculated values of heterogeneity metric associated to the hyperparameters used as key.
The Machine Learning Engine: H2O.ai
As the causal inference adjustment requires a hyperparameter search not once, but for each iteration of the inverse probability weight process until it converges, we need to select a ML engine very fast and efficient in resource consumption. Giving preference for the ones that would allow the execution of this procedure in multi-core laptops or clusters without changing too much the code implemented.
We think that H2O.ai is such engine. As its description in its websites says:
“H2O is the world’s leading open source machine learning platform. H2O is used by over 60,000 data scientists and more than 7,000 organizations around the world. H2O is an open source, in-memory, distributed, fast, and scalable machine learning and predictive analytics platform that allows you to build machine learning models on big data and provides easy productionalization of those models in an enterprise environment. H2O’s core code is written in Java. Inside H2O, a Distributed Key/Value store is used to access and reference data, models, objects, etc., across all nodes and machines. The algorithms are implemented on top of H2O’s distributed Map/Reduce framework and utilize the Java Fork/Join framework for multi-threading. The data is read in parallel and is distributed across the cluster and stored in memory in a columnar format in a compressed way. H2O’s data parser has built-in intelligence to guess the schema of the incoming dataset and supports data ingest from multiple sources in various formats. H2O’s REST API allows access to all the capabilities of H2O from an external program or script via JSON over HTTP. The Rest API is used by H2O’s web interface (Flow UI), R binding (H2O-R), and Python binding (H2O-Python). The speed, quality, ease-of-use, and model-deployment for the various cutting edge Supervised and Unsupervised algorithms like Deep Learning, Tree Ensembles, and GLRM make H2O a highly sought after API for big data data science.”
Wrapping up
Now we have all elements to implement our causal inference algorithm in R. In the next post we’ll show some demos with source code to test how our approach performs in different setups and with different machine learning algorithms.
Previous Post – Causal Inference Theory With Observational Data = Structural Causal Model + Propensity Score + Iterative Inverse Probability Weighting
Any feedback send to frederico.nogueira@gmail.com